You are here: vision-research.eu » Vision Research » Visionary of the Quarter » Sandro Banfi (Q01-2017)
The role of noncoding RNAs in ocular development and function

The research topics of Sandri Banfi
In the past decade, regulatory noncoding RNAs (ncRNAs) have emerged as fundamental components of the mammalian transcriptome. They are considered crucial effectors in a variety of biological processes, both in physiological and pathological conditions. The most important subclasses are constituted by microRNAs (miRNAs) and by long noncoding RNAs (lncRNAs). MicroRNAs are small, endogenous RNAs that negatively regulate gene expression post-transcriptionally by binding to target sites in the 3’ untranslated region (UTR) of messenger RNAs. LncRNAs represent a more heterogeneous class of noncoding transcripts that can control different cellular programs at either the transcriptional or post-transcriptional levels.
A major focus of our research is the dissection of the role of regulatory ncRNAs in ocular development and visual function, with an emphasis on miRNAs. Towards this goal, we rely on the combination of functional genomics, bioinformatics and molecular biology approaches. Our main findings in this field are summarized below.
1.) Functional genomics and in silico assessment of miRNA contribution to retinal function
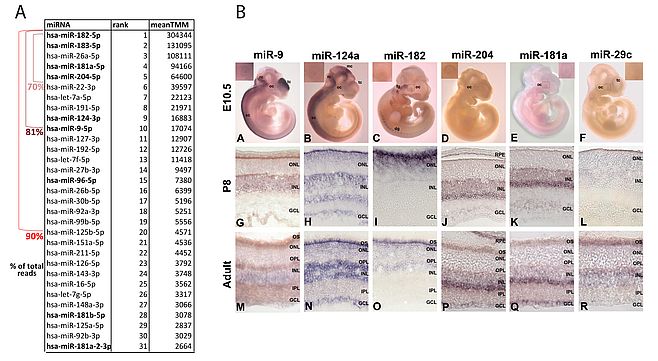
We characterized at a global level the expression pattern of miRNAs in the mammalian retina (both in mouse and in human) using microarray1 and subsequently Next Generation Sequencing procedures2. We coupled this analysis with a detailed characterization of the spatiotemporal distribution in the eye and retina of selected miRNAs by RNA in situ hybridization3.This work has allowed us to generate the most comprehensive thus far miRNA transcriptome atlas (miRNome) of the human retina and to gain insight into the catalog of the miRNAs expressed at the most significant levels in this tissue (Figure 1). We also analysed the complexity of the retinal miRNome at a nucleotide resolution by characterizing the alternative sequence variants of known miRNAs (isomiRs) and defining their contribution to the retinal miRNome (http://miretina.tigem.it)2. In parallel, we generated a comprehensive atlas of mRNA expression and co-regulation in the human retina4. The combined analysis of the miRNA and of the mRNA transcriptome atlases of the human retina represents a powerful resource for the definition of the main transcriptional pathways underlying retinal function.
Understanding miRNA function, requires the reliable prediction of target genes. We decided to apply co-expression-based procedures to devise effective algorithms for miRNA function predictions. We first developed the HocTAR tool, which recognized the targets of intragenic microRNAs by combining the expression analysis of their host genes and sequence prediction softwares (http://hoctar.tigem.it)5. More recently, we further exploited the integration of co-expression data analysis and of target prediction and implemented the co-expression Meta-analysis of miRNA Targets (CoMeTa) tool to gain insight into the biological functions of all miRNAs and to provide a comprehensive, genome-wide scale analysis of human miRNA regulatory networks6. Finally, we also generated a database (CoGemiR) specific for the assessment of the genomic organization of miRNAs across evolution (http://cogemir.tigem.it)7.
2.) Determination of the role of specific ncRNAs in retinal function in health and disease
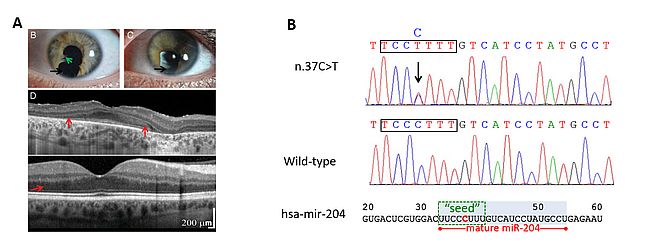
In the past few years, using a variety of both in vitro and in vivo approaches, we have gained insight into the fundamental role of the miR-204 in several aspects of eye development and function (Figure 2). In particular, we found that this miRNA plays a role in lens formation and in the establishment of a proper dorsal-ventral retinal axis8 as well as in retinal axon guidance9. More recently, we also discovered that miR-204 is necessary for proper photoreceptor function as revealed by the finding that a point mutation in its mature sequence plays a pathogenic role in an autosomal dominantly inherited condition of syndromic retinal dystrophy10. The latter represents not only the first example of a microRNA mutation with a causative role in inherited retinal dystrophies in patients but also the first instance of a microRNA-caused genetic disease most likely occurring via a gain-of-function mechanism. Overall, our findings indicate that miR-204 is a new putative “master regulator” of eye development and function and support the notion that miRNAs can represent primary pathogenic agents in human retinal disorders.
We have also been involved in the identification and characterization of lncRNAs with a presumably relevant role in retinal function. We have identified, and begun to characterize, a subset of lncRNAs associated with transcription factors that are known to play a basic role in retinal development in vertebrates11, 12. We demonstrated that one of them, i.e., Vax2os1, is involved in the control of cell cycle progression of photoreceptor progenitor cells in the ventral retina13.
The following are additional research interests in our laboratory:
- Dissection of the molecular basis of inherited eye disorders
- Study of the molecular mechanisms underlying retinal development
- Design of advanced therapeutic strategies for inherited retinal disorders
Bibliography
- Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, Dolle P, Banfi S (2010). miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics 11 715.
- Karali M, Persico M, Mutarelli M, Carissimo A, Pizzo M, Singh Marwah V, Ambrosio C, Pinelli M, Carrella D, Ferrari S, Ponzin D, Nigro V, di Bernardo D, Banfi S (2016). High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res 44 (4):1525-1540.
- Karali M, Peluso I, Marigo V, Banfi S (2007). Identification and characterization of microRNAs expressed in the mouse eye. Invest Ophthalmol Vis Sci 48 (2):509-515.
- Pinelli M, Carissimo A, Cutillo L, Lai CH, Mutarelli M, Moretti MN, Singh MV, Karali M, Carrella D, Pizzo M, Russo F, Ferrari S, Ponzin D, Angelini C, Banfi S, di Bernardo D (2016). An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res.
- Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S (2008). MicroRNA target prediction by expression analysis of host genes. Genome Res.
- Gennarino VA, D'Angelo G, Dharmalingam G, Fernandez S, Russolillo G, Sanges R, Mutarelli M, Belcastro V, Ballabio A, Verde P, Sardiello M, Banfi S (2012). Identification of microRNA-regulated gene networks by expression analysis of target genes. Genome Res.
- Maselli V, Di Bernardo D, Banfi S (2008). CoGemiR: a comparative genomics microRNA database. BMC Genomics 9 457.
- Conte I, Carrella S, Avellino R, Karali M, Marco-Ferreres R, Bovolenta P, Banfi S (2010). miR-204 is required for lens and retinal development via Meis2 targeting. Proc Natl Acad Sci U S A 107 (35):15491-15496.
- Conte I, Merella S, Garcia-Manteiga JM, Migliore C, Lazarevic D, Carrella S, Marco-Ferreres R, Avellino R, Davidson NP, Emmett W, Sanges R, Bockett N, Van Heel D, Meroni G, Bovolenta P, Stupka E, Banfi S (2014). The combination of transcriptomics and informatics identifies pathways targeted by miR-204 during neurogenesis and axon guidance. Nucleic Acids Res 42 (12):7793-7806.
- Conte I, Hadfield KD, Barbato S, Carrella S, Pizzo M, Bhat RS, Carissimo A, Karali M, Porter LF, Urquhart J, Hateley S, O'Sullivan J, Manson FD, Neuhauss SC, Banfi S, Black GC (2015). MiR-204 is responsible for inherited retinal dystrophy associated with ocular coloboma. Proc Natl Acad Sci U S A 112 (25):E3236-3245.
- Alfano G, Vitiello C, Caccioppoli C, Caramico T, Carola A, Szego MJ, McInnes RR, Auricchio A, Banfi S (2005). Natural antisense transcripts associated with genes involved in eye development. Hum Mol Genet 14 (7):913-923.
- Alfano G, Conte I, Caramico T, Avellino R, Arno B, Pizzo MT, Tanimoto N, Beck SC, Huber G, Dolle P, Seeliger MW, Banfi S (2011). Vax2 regulates retinoic acid distribution and cone opsin expression in the vertebrate eye. Development 138 (2):261-271.
- Meola N, Pizzo M, Alfano G, Surace EM, Banfi S (2012). The long noncoding RNA Vax2os1 controls the cell cycle progression of photoreceptor progenitors in the mouse retina. RNA 18 (1):111-123.
Contact
Prof. Sandro Banfi
Mail:
Department of Biochemistry, Biophysics and General Pathology
Universita’ degli studi della Campania “Luigi Vanvitelli"
via Luigi De Crecchio 7 - 80138 Naples Italy
and
Telethon Institute of Genetics and Medicine, Pozzuoli
Via Campi Flegrei, 34
80078 Pozzuoli (NA), Italy
Phone and E-mail
Tel: +39 08119230628
Fax +39-081-19230651
E-mail: banfi[at]tigem.it