You are here: vision-research.eu » Vision Research » Visionary of the Quarter » Herwig Baier (Q02-2021)
The research work of Professor Herwig Baier
 |
Overview of our research questions
My laboratory is exploring how genetic information assembles neuronal circuits and how these circuits in turn control behavior. The fantastic people in my group, who drive this field forward, are portrayed in Fig. 1. Our main focus has been the zebrafish visual system, although we have recently branched out into other species and other parts of the brain. Zebrafish have become a premier research model for behavioral genetics, sensory and systems neuroscience, and neuroethology.
Our current interests can be summarized by the following bullet points:
- Functional dissection of visual and visuomotor pathways at the cellular and synaptic scale.
- Neural basis of elementary cognitive processes, including decision-making, attention, memory and social affiliation.
- Machine learning-supported, kinematic analysis of behavior.
- Linking circuit architecture to behavioral adaptations.
- Innovation, improvement and adaptation of optical, genetic and optogenetic methods.
- Development of a multimodal, cellular-resolution atlas of the zebrafish brain.
Over the years, we have made contributions to each of these areas of investigation. Here I will summarize some of our advances in neural circuit analysis, technology development and the Max Planck Zebrafish Brain Atlas.
Neural pathways for visually guided behaviors
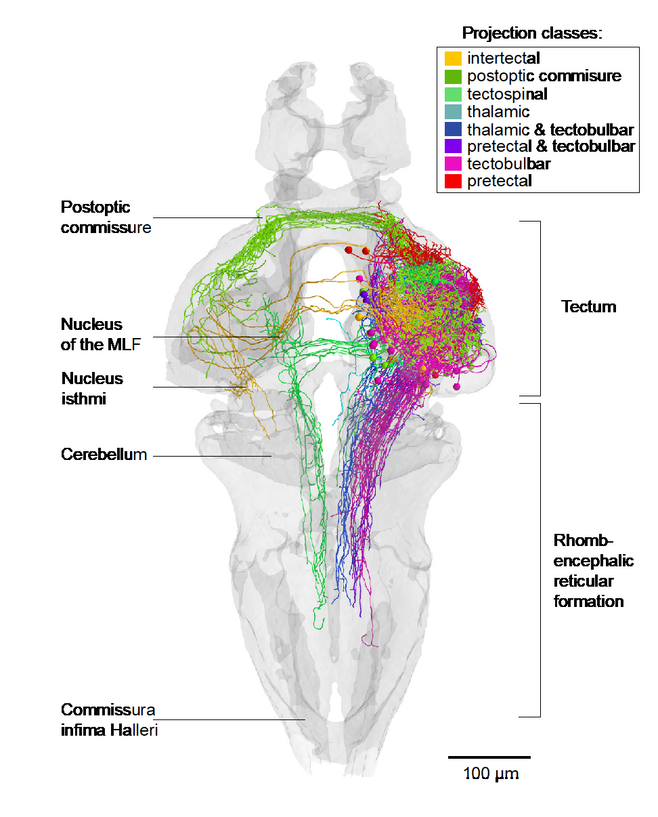
Over the past decade, we have identified neural pathways and circuits that convert sensory input into motor output for select behaviors, such as phototaxis, prey capture, looming-evoked escape, optokinetic and optomotor responses and the visual recognition of conspecifics. For several of these behaviors, our group has discovered the pathways connecting the retina to the tectum, pretectum, thalamus or preoptic area (Semmelhack et al., 2014; Temizer et al., 2015; Wu et al., 2020; Kölsch et al., 2020). We have also succeeded in mapping downstream connectivity to premotor areas in the tegmentum and reticular formation (e. g., Fig. 2; Helmbrecht et al., 2018; Kramer et al., 2019). This body of work has led to the general hypothesis that stimulus quality (e. g., optic flow vs. prey-like vs. threat vs. conspecific) and valence (attractive vs. aversive vs. neutral) are encoded by anatomically segregated circuits, which receive inputs from staggered arrays of matched filters. These dedicated pathways mutually suppress each other through reciprocal inhibitory connections (see, e. g., Barker and Baier, 2015; Filosa et al., 2016). Such a canonical wiring principle seems to underlie the diverse circuit architectures serving stimulus selection, object identification and behavioral decisions.
Feature-selective retinal ganglion cells (RGCs) represent prime examples of matched filters. To enable genetic access to specific visual processing streams, our group has molecularly catalogued 33 RGC types in larva and adult zebrafish (Kölsch et al., 2021). This was done by single-cell RNA sequencing (scRNA-seq; initially in collaboration with Josh Sanes and Karthik Shekhar, Harvard University and the Broad Institute). Fig. 3 shows schematically five transgenic lines, which were generated using CRISPR/Cas9 knock-ins of type-specific marker genes. Each of the lines provides experimental access to a small subpopulation of RGCs. Novel intersectional genetic strategies, combining QF2 or Gal4, and Cre recombination, are allowing us now to target single RGC types for recording or manipulation (Kölsch et al., 2020). Utilizing this resource and building on the laboratory’s previous work (Robles et al., 2013; 2014), we discovered a pathway serving phototaxis, which originates in an RGC subclass that express the transcription factor Eomesa. Based on a morphological survey of RGC dendrite stratification and axonal projection patterns (Robles et al., 2014), I suspect the total number of RGC types will be greater than the 30, or so, molecular classes, maybe double as many.
Advances in imaging, optogenetics and computer science
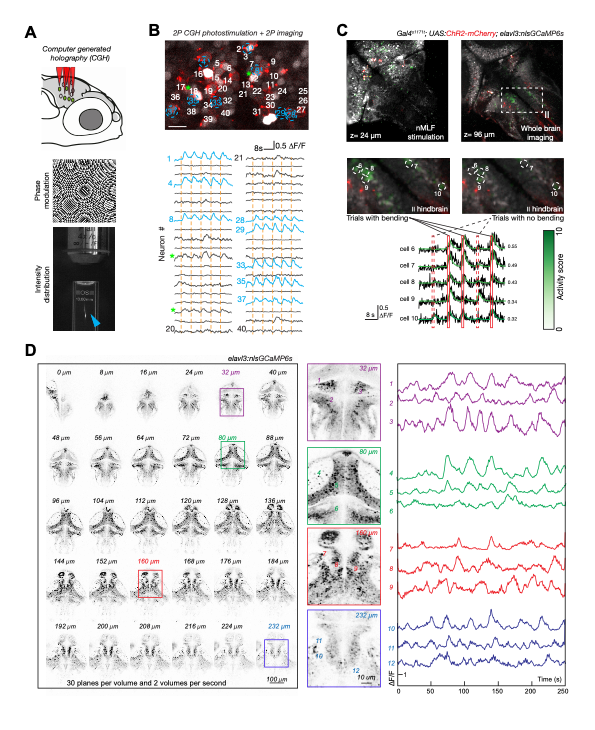
Genetic access and transparency of larval zebrafish makes them an excellent model for functional investigation of sensorimotor circuits. In order to fully leverage this advantage, our group develops advanced optical capabilities for recording and manipulating neuron activity. Two-photon calcium imaging is a common technique in the lab; however, standard two-photon microscopes can only record from a single plane, missing relevant correlations between neurons. We have therefore devised rapid volumetric imaging methods, which enable recordings from multiple Z planes (see Fig. 4).
Imaging provides correlative measurements and is often incapable of providing causal evidence or validating proposed circuit mechanisms. Therefore, our group has been advancing an approach based on spatially precise, two-photon computer-generated holographic optogenetics for manipulating activity at the scale of individual neurons (Dal Maschio et al., 2017). A new system built in the lab features optics with improved field of view and resolution for flexible imaging of multiple subvolumes, as well as improvements to the detection pathway and software (Fig. 4). The combined abilities of such an instrument are enabling new kinds of experiments, such as photostimulating multiple, functionally defined neurons, while imaging in downstream areas to uncover causal flows of circuit activity.
Max Planck Zebrafish Brain Atlas
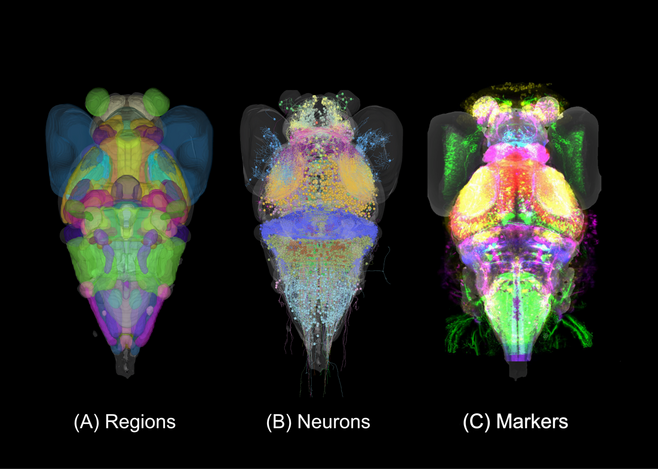
Lack of neuroanatomical knowledge hampers progress in systems neuroscience. We have therefore assembled a cellular-resolution atlas of the larval zebrafish brain, which supports not only our own research, but also provides a resource to the wider community (Kunst et al., 2019). The Max Planck Zebrafish Brain Atlas, in its latest version, integrates 116 expert-annotated brain regions, more than 4,000 single neuron morphologies and a growing number (>450) of transgenic lines. These data modalities are registered to the same standard brain space (Fig. 5).
The atlas portal on the website (https://fishatlas.neuro.mpg.de) offers refined analysis tools and also the possibility of uploading, visualization and downloading of data. Several recent papers have relied heavily on this resource (Helmbrecht et al., 2018; Kramer et al., 2019; Wu et al., 2020; Pantoja et al., 2020; Förster et al., 2020). An improved version (working title “mapZeBrain 2.0”) will integrate additional data modalities, such as functional imaging and gene expression data, offer an API for related efforts in other systems, and will sport an improved front-end user interface. The atlas attracts a rapidly growing number of external users (>2,500 unique page views as of April 2021).
Vision-related publications (selection)
- Barker AJ, Helmbrecht TO, Grob AA, Baier H (2021) Functional, molecular and morphological heterogeneity of superficial interneurons in the larval zebrafish tectum. J Comp Neurol doi: 10.1002/cne.25082.
- Fernandes AM, Mearns DS, Donovan JC, Larsch J, Helmbrecht TO, Kölsch Y, Laurell E, Kawakami K, Dal Maschio M, Baier H (2021). Neural circuitry for stimulus selection in the zebrafish visual system. Neuron 109: 805-822.
- Kölsch Y, Hahn J, Sappington A, Stemmer M, Fernandes AM, Helmbrecht TO, Lele S, Butrus S, Laurell E, Arnold-Ammer I, Shekhar K, Sanes JR, Baier H (2021). Molecular classification of zebrafish retinal ganglion cells links genes to cell types to behavior. Neuron 109:645-662.
- Förster D, Helmbrecht TO, Mearns DS, Jordan L, Mokayes N, Baier H (2020). Retinotectal circuitry of larval zebrafish is adapted to detection and pursuit of prey. Elife 9, e58596.
- Wu Y, Dal Maschio M, Kubo F, Baier H (2020). An optical illusion pinpoints an essential circuit node for global motion processing. Neuron 108: 1-13.
- Kramer A, Wu Y, Baier H, Kubo F (2019). Neuronal architecture of a visual center that processes optic flow. Neuron 103: 118-132.
- Kunst M, Laurell E, Mokayes N, Kramer A, Kubo F, Fernandes AM, Förster D, Dal Maschio M, Baier H (2019). A cellular-resolution atlas of the larval zebrafish brain. Neuron 103: 21-38.
- Helmbrecht TO, dal Maschio M, Donovan JC, Koutsouli S, Baier H (2018). Topography of a visuomotor transformation. Neuron 100: 1429-1445.
- Larsch J, Baier H (2018). Biological motion as an innate perceptual mechanism driving social affiliation. Current Biology 28: 3523-3532.
- Förster D, Dal Maschio M, Laurell E, Baier H (2017). An optogenetic toolbox for unbiased discovery of functionally connected cells in neural circuits. Nature Communications 8: 116.
- Dal Maschio M, Donovan JC, Helmbrecht TO, Baier H (2017). Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron 94: 774-789.
- Filosa A, Barker A, Dal Maschio M, Baier H (2016). Feeding state modulates behavioral choice and processing of prey stimuli in the zebrafish tectum. Neuron 90: 596-608.
- Barker A, Baier H (2015). Sensorimotor decision-making in the zebrafish tectum. Current Biology 25: 2804-14.
- Temizer I, Donovan JC, Baier H, Semmelhack JL (2015). A visual pathway for looming-evoked escape in larval zebrafish. Current Biology 25: 1823-34.
- Semmelhack JL, Donovan JC, Thiele TR, Kuehn E, Laurell E, Baier H (2014). A dedicated visual pathway for prey detection in larval zebrafish. eLife 10.7554.
- Robles E. Laurell E, Baier H (2014). The retinal projectome reveals brain-area-specific visual representations generated by ganglion cell diversity. Current Biology 24: 2085-2096.
- Thiele T, Donovan JC, Baier H (2014). Descending control of swim posture by a midbrain nucleus in zebrafish. Neuron 83: 679-691.
- Kubo F, Hablitzel B, Dal Maschio M, Driever W, Baier H, Arrenberg AB (2014). Functional architecture of an optic flow responsive area that drives horizontal eye movements in zebrafish. Neuron 81:1344-59.
- Baier H (2013). Synaptic laminae in the visual system – Molecular mechanisms forming layers of perception. Ann Rev Cell Dev Biol 29:385-416.
- Robles E, Filosa A, Baier H (2013). Precise lamination of retinal axons generates multiple parallel input pathways in the tectum. J Neurosci 33:5027-5039.
- Xiao T, Staub W, Robles E, Gosse NJ, Cole GJ, Baier H (2011). Assembly of lamina-specific neuronal connections by Slit bound to type IV collagen. Cell 146:164-176.
- Huberman AD, Clandinin TR, Baier H (2010). Molecular and cellular mechanisms of lamina-specific axon targeting. Cold Spring Harb Perspect Biol 2:a001743.
- Del Bene F, Wyart C, Robles E, Tran A, Looger L, Scott EK, Isacoff EY, Baier H (2010). Filtering of visual information in the tectum by an identified neural circuit. Science 330:669-673.
- Baier H, Scott EK (2009). Genetic and optical targeting of neural circuits and behavior – zebrafish in the spotlight. Curr Opin Neurobiol 19:553-560.
- Wyart C, Del Bene F, Warp E, Scott EK, Trauner D, Baier H, Isacoff EY (2009). Optogenetic dissection of a behavioral module in the vertebrate spinal cord. Nature 461:407-410.
- Gosse NJ, Nevin LM, Baier H (2008). Retinotopic order in the absence of axon competition. Nature 452:892-895.
- Del Bene F, Wehman A, Link BA, Baier H (2008). Regulation of neurogenesis by interkinetic nuclear migration through an apical-basal notch gradient. Cell 134:1055-1065.
- Sumbre G, Muto A, Baier H, Poo MM (2008). Entrained rhythmic activities of neuronal ensembles as perceptual memory of time interval. Nature 456:102-106.
- Smear MC, Tao HW, Staub W, Orger MB, Gosse NJ, Liu Y, Takahashi KD, Poo MM, Baier H (2007). Vesicular glutamate transport at a central synapse limits the the acuity of visual perception in zebrafish. Neuron 53:65-77.
- Scott EK, Mason L, Arrenberg A, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H (2007). Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nature Methods 4:323-326.
- Hua Y, Smear MC, Baier H, Smith SJ (2005). Activity-based competition regulates axon growth in vivo. Nature 434:1022-1026
- Page-McCaw P, Chung SC, Muto A, Roeser T, Staub W, Finger-Baier KC, Korenbrot JI, Baier H (2004). Tyrosinase is required for retinal network adaptation to bright light. Nature Neuroscience 7:1329-1336.
- Orger MB, Smear MC, Anstis SM, Baier H (2000). Perception of Fourier and non-Fourier motion by larval zebrafish. Nature Neuroscience 3:1128–1133.
- Neuhauss SCF., Biehlmaier O, Seeliger MW, Das T, Kohler K, Harris WA, Baier H (1999). Genetic disorders of vision revealed by a behavioral screen of four-hundred essential loci in zebrafish. J Neurosci 19:8603-8615.

Professor Dr. Herwig Baier
Director of the Department Genes–Circuits–Behavior
Max Planck Institute of Neurobiology
Am Klopferspitz 18
82152 Martinsried near Munich
Phone: +49-89-8578-3200
Email: hbaier[at]neuro.mpg.de
Website: https://www.neuro.mpg.de/baier